Simmerling lab research on COVID-19
(Click the “research” tab above to see our web page on the rest of our research projects)
This site is not meant to be a medical resource for COVID-19 or illnesses caused by coronaviruses. Please refer to The Centers for Disease Control and Prevention (CDC ) site.
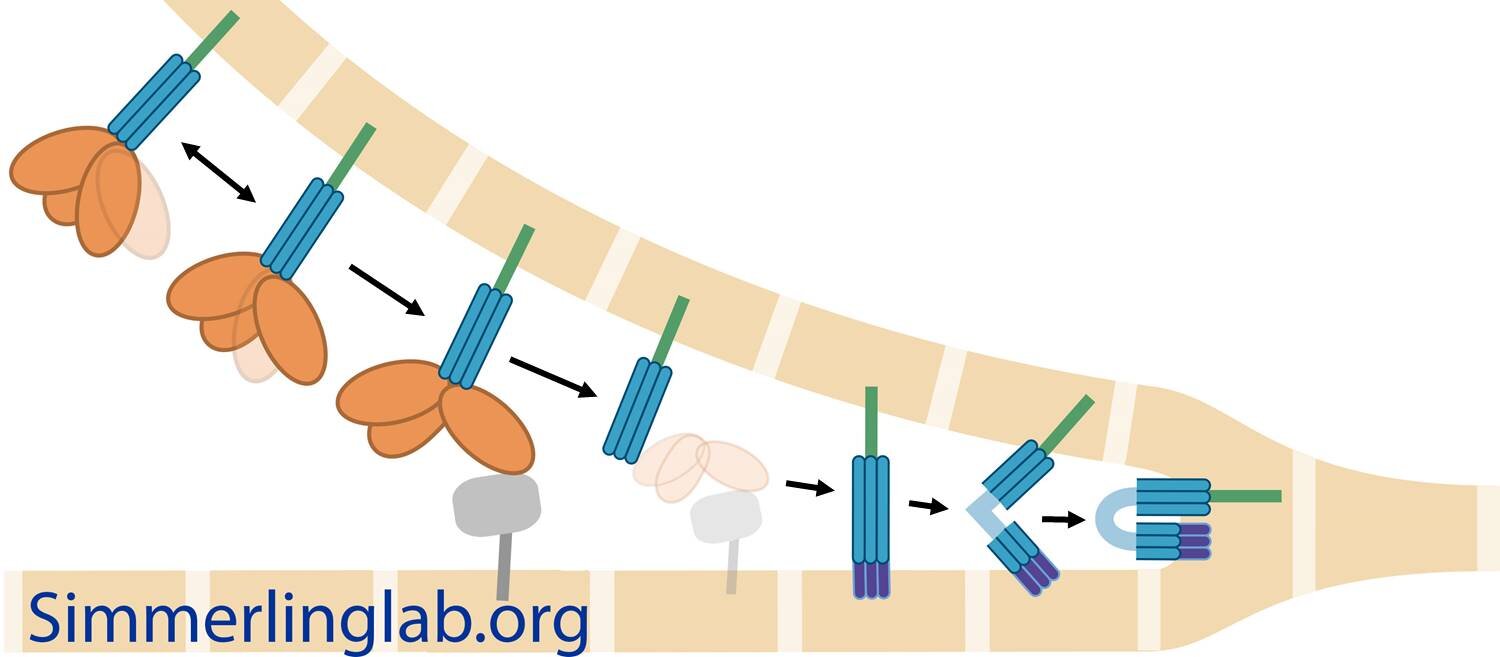
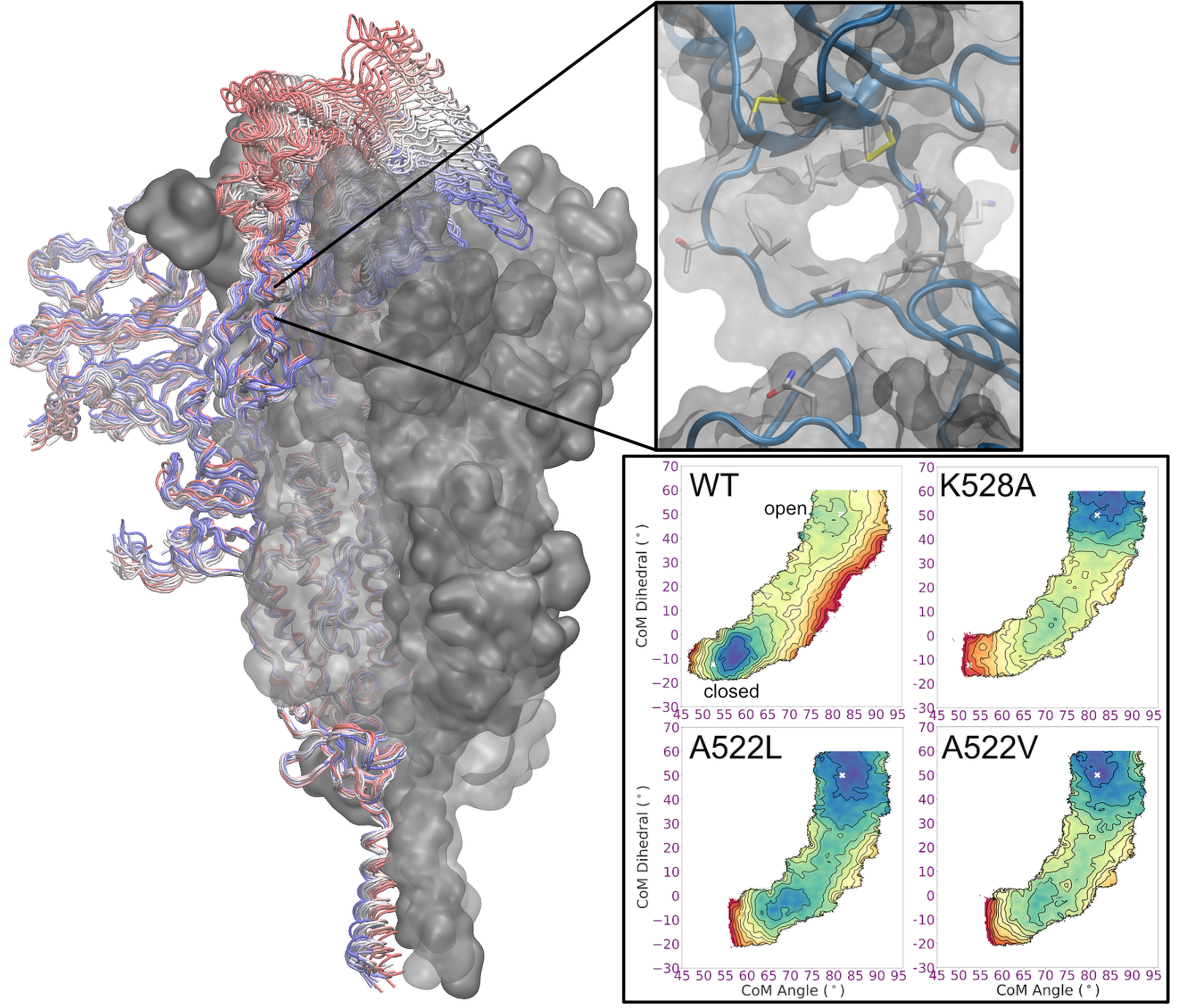
Coronaviruses have spike glycoproteins decorating their surface; the virus uses the spikes to gain entry into host cells. Experiments have revealed the structure of the spike before and after viral entry, and dramatic changes take place. We aim to understand the full details of these conformational changes, including understanding what triggers the process, each of the steps that occurs along the way, and whether interfering with this structural change could provide an avenue for blocking coronaviruses from infecting host cells.